eDNA metabarcoding
I use envrionmental DNA metabarcoding as tool to explore the entire soil diversity including plant roots.
DNA extraction and amplification
Using primers for prokaryotes (16S) as well as eukaryotes (18S) and specifically fungi (ITS and LSU), plants (rbcL), animals, insects, arthropods.
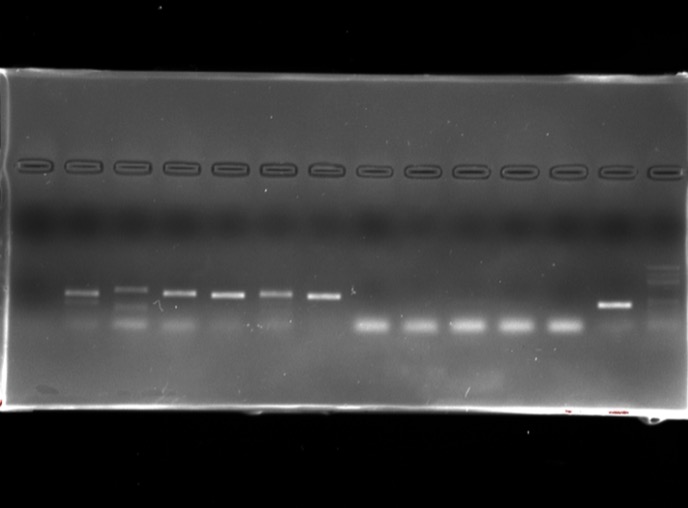
Bioinformatics
I have been using several pipelines in order to apply bioinformatic treatments to raw sequence data such as DADA2 and OBITools that are published online and available on my GitHub but also as tutorials.
See for example here a custom pipeline with fungal sequences I obtained using Illumina MiSeq sequencing platform from soil samples.
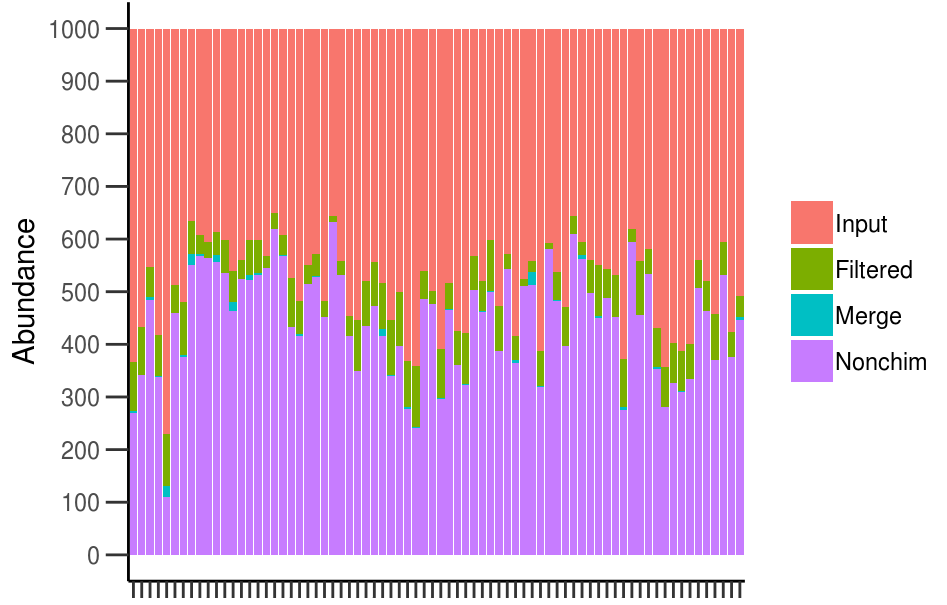
The figure shows the percentage of reads per samples kept after different steps of “denoising”.
Analyses of sequencing data
DNA sequencing can generate large amount of data that can be analyzed using specific tools such as Phyloseq that allow to handle such community data in a reproducible and accessible way.
See the full project on eDNA metabarcoding: From raw data to RDA (“fun” tutorials on DADA2 and phyloseq available here).
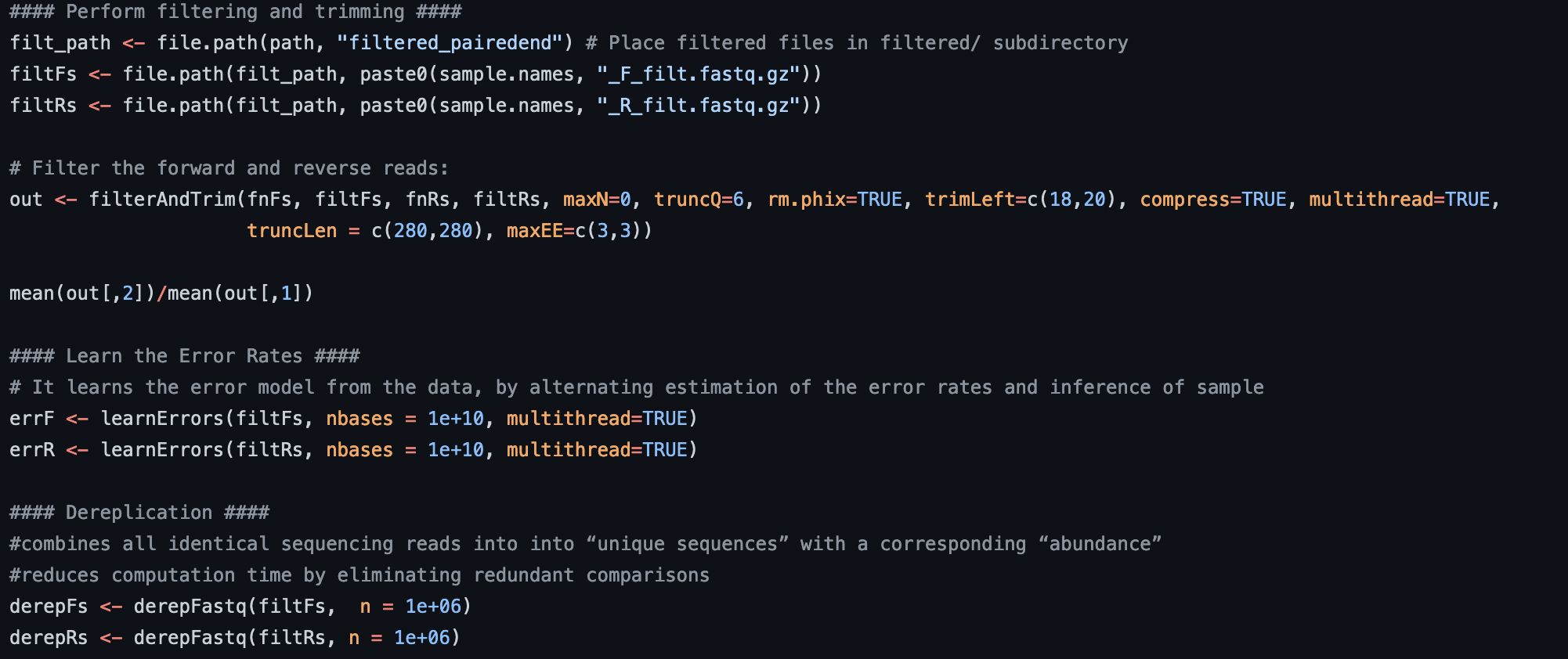 Example of bioinformatic code. Full code here.
Example of bioinformatic code. Full code here.
I also organized workshops, with the help of colleagues, to train the next generation of scientists to use metabarcoding approaches. All the materials of the BIOME workshop on advanced bioinformatics can be found here.
 BIOME workshop participants, Laurentides
BIOME workshop participants, Laurentides
 Teaching bioinformatics and biostatistics
Teaching bioinformatics and biostatistics